|
miRGator v3.0
a microRNA portal in deep sequencing era
|
||
|
About miRGator
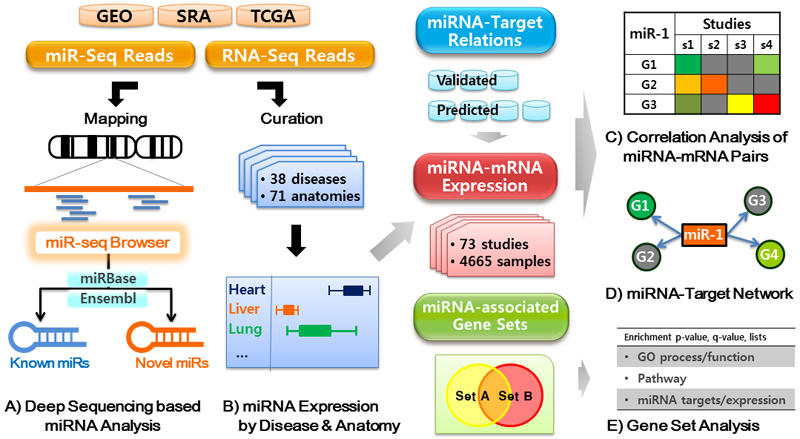
miRGator aims to be the microRNA (miRNA) portal encompassing microRNA diversity, expression profiles, target relationships, and various supporting tools. Major imporvement in this update is the inclusion of
deep sequencing data, which became the principal resources on microRNA diversity and expression.
We have collected 73 deep sequencing datasets on human samples from GEO, SRA, and TCGA archives, which amounts to 4.1 billion short reads and 2.5 billion aligned reads.
We have curated these data into 38 diseases and 71 anatomic categories. The schematic for miRGator v3.0 is shown below.
|
||
|
Major Features
1) miR-seq browser :
miR-seq browser that visualizes the sequence alignment and normalized read counts with the secondary structure information simultaneously in concurrent windows. Short reads related to iso-miRs and miRNA editing can be readily identified with the corresponding expression values (read counts) in multiple samples. This helps users to easily examine the relationships between the structure of precursor, the sequences and abundance of final products, and thereby will facilitate the studies on miRNA biogenesis and regulation.
2) miRNA catalogues and Expression profiles :
miRNA catalogues and Expression profiles in various diseases, organs, and tissues based on deep sequencing data, which would provide an unbiased catalog of miRNA repertoire and accurate estimation of abundance. Our computational pipeline identified 1856 mature miRNAs and 6424 ncRNAs, and mirDeep2 predicted 508 mature and 304 pre-miRNAs. We also provide the list of differentially expressed miRNAs where normal samples are available.
3) miRNA-mRNA target relations and expression correlations :
miRNA-mRNA target relations and expression correlations integrated for identifying reliable targets. We have merged 3 databases of validated targets and 6 databases of predicted targets. Inverse correlation of gene expression is a strong evidence of genuine targetedness, and we have specifically collected studies with miRNA and mRNA expression data from the same samples. The result is visually represented in two formats of correlation heat map and network views.
|
||
|
References
Sooyoung Cho, Insu Jang, Yukyung Jun, Suhyeon Yoon, Minjeong Ko, Yeajee Kwon, Ikjung Choi, Hyeshik Chang, Daeun Ryu, Byungwook Lee, V. Narry Kim, Wankyu Kim, Sanghyuk Lee
Nucleic Acids Res. 2013 Jan;41(Database issue):D252-57.
Sooyoung Cho, Yukyung Jun, Sanghyun Lee, Hyung-Seok Choi, Sungchul Jung, Youngjun Jang, Charny Park, Sangok Kim, Sanghyuk Lee, and Wankyu Kim
Nucleic Acids Res. 2011 Jan;39(Database issue):D158-62.
Seungyoon Nam, Bumjin Kim, Seokmin Shin and Sanghyuk Lee
Nucleic Acids Res. 2008 Jan;36(Database issue):D159-64. |

